REVNANO
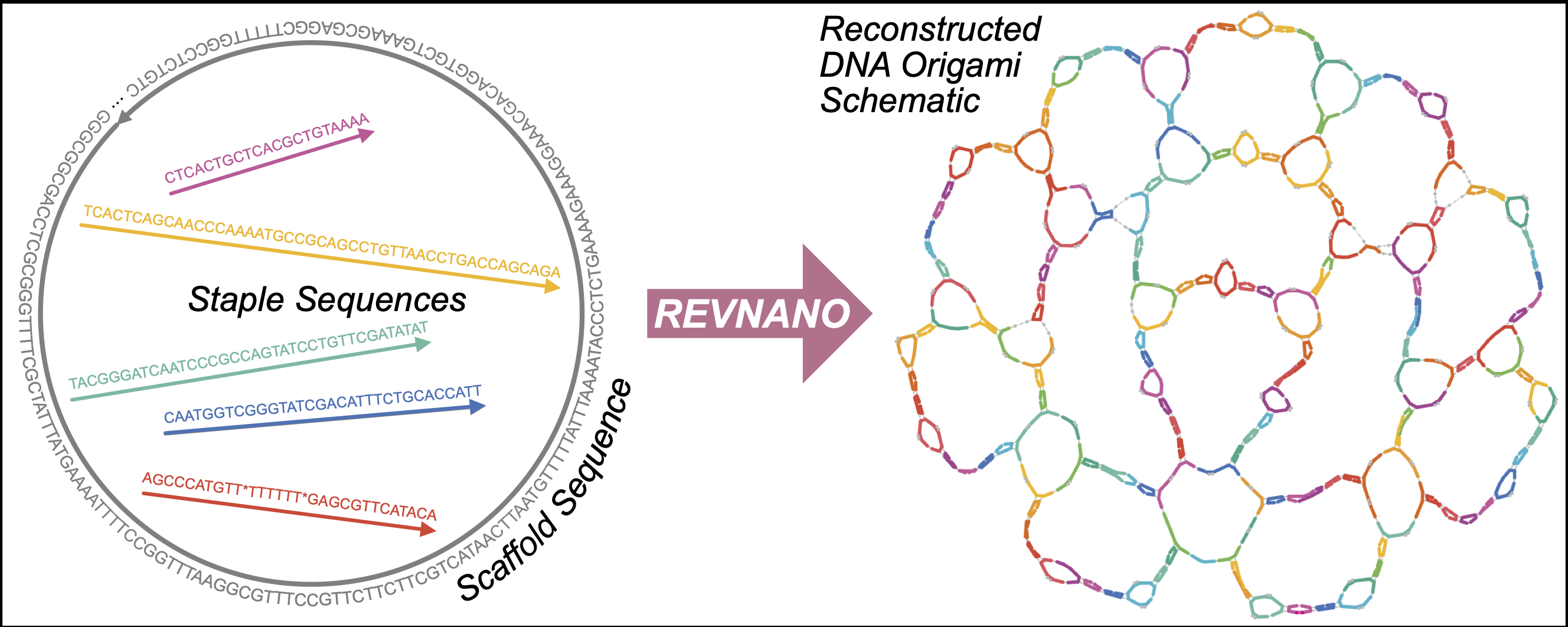
REVNANO is a computational tool that reverse engineers a schematic diagram for a nucleic acid origami nanostructure, given just the the raw scaffold and staple sequences for that nanostructure. Written in Python, the code is available as an open source repository on Bitbucket under a MIT Licence.
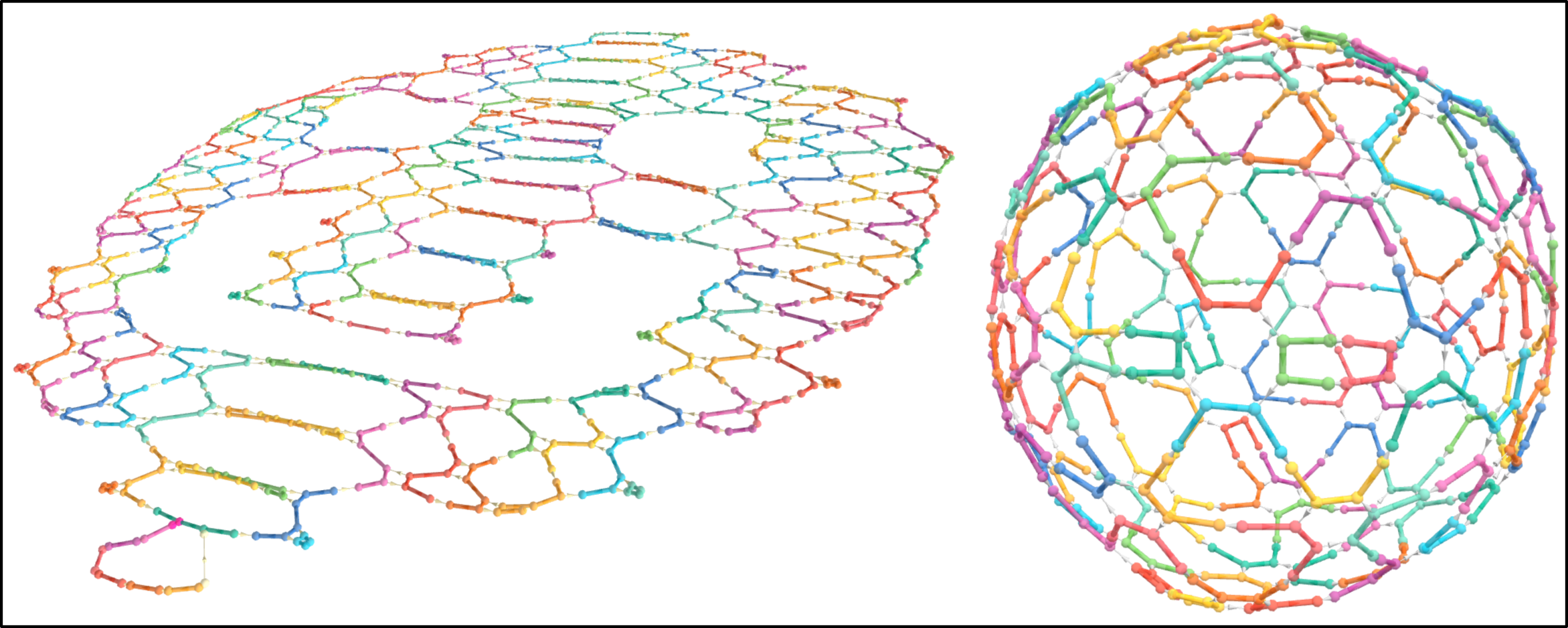
REVNANO works with DNA origami shapes, RNA origami shapes or hybrid origami shapes (e.g. RNA scaffold, DNA staples) in both 2-dimensions and 3-dimensions.
Input to the solver is a simple plain text sequence file like this, and output is an interactive HTML page showing an approximate ‘guide schematic’ of the origami nanostructure like this.
While all current software for DNA origami nanostructure design implements the forward problem of converting a geometric schematic to sequences, REVNANO solves the inverse of this problem.
Reverse engineering an (approximate) origami guide schematic from scaffold and staple sequences is generally useful for the following reasons:
Most DNA origami designs are published as raw sequences only in supplementary materials: this is the de facto standard of origami design exchange. Typically, no electronic origami design files are provided. The interactive guide schematic produced from sequences can assist in entering a DNA origami design back into a DNA origami CAD tool, hence recovering an electronic schematic file for the origami
The guide schematic allows examination of features of the origami in greater detail than is typically provided in a publication
Reversing sequences back to a guide schematic can provide a double-check that there are no sequence errors in the sequence lists
If a publication is vague about scaffold sequence, the staple set provided can be reversed against different scaffold sequences until the correct scaffold that makes the origami is found
Ease of reversing sequences back to a guide schematic is likely to be crudely correlated with lack of kinetic traps in the physical self-assembly of an origami nanostructure
REVNANO is written in Python and can be run from the command line or as part of a JupyterLab notebook on a single computer. Typical running times on a modern single computer ranges from a few seconds, up to around 5 minutes for larger origami designs based on e.g. M13 scaffold.
The REVNANO solver is described in more detail and benchmarked on a test set of 36 origamis in our 2023 paper in Computational and Structural Biotechnology Journal. If REVNANO is useful in your work, please consider citing our paper. See How to Cite Our Paper.
Finally, REVNANO makes use of a newly developed Nucleic Acid Origami Contact Map Format which is worth a look in its own right, as it has many useful features for creating, converting and modifying origami staple-scaffold contact maps.
Below is described how to install and run REVNANO. Also check out the Paper Examples page where there are sequence input files for all 36 origamis in our paper, to test out for yourself.